|
Overview
of the EpiFOS Fosmid Library Production Process
Fosmid
vectors 1-3 provide an improved method for cloning and the
stable maintenance of cosmid- sized (35-45kb) libraries in
E. coli . The stability of such large constructs
in vivo is facilitated by the pEpiFOS TM -5 vector that maintains
the clones at single copy in the cell.
The EpiFOS Fosmid Library Production Kit will produce a complete
and unbiased primary fosmid library. The kit utilizes a novel
strategy of cloning randomly sheared, end-repaired DNA. Shearing
the DNA leads to the generation of highly random DNA fragments
in contrast to more biased libraries that result from fragmenting
the DNA by partial restriction digests.
The
steps involved (protocols for steps 2-7 are included in this
manual):
I.
Purify DNA from the desired source (the kit dues not supply
materials for this step).
2
. Shear the DNA to approximately 40 Kb fragments.
3.
End-repair the sheared DNA to blunt, 5'-phosphorylated ends.
4.
Size-resolve the end-repaired DNA by Low Melting Point
(LMP) agarose gel electrophoresis.
5.
Purify the blunt-end DNA from the LMP agarose gel.
6.
Ligate the blunt-end DNA to the Cloning-Ready pEpiFOS-5 vector.
7.
Package the ligated DNA and plate on EPI100 TM plating cells.
Grow clones overnight.
Fosmid
Vector Details
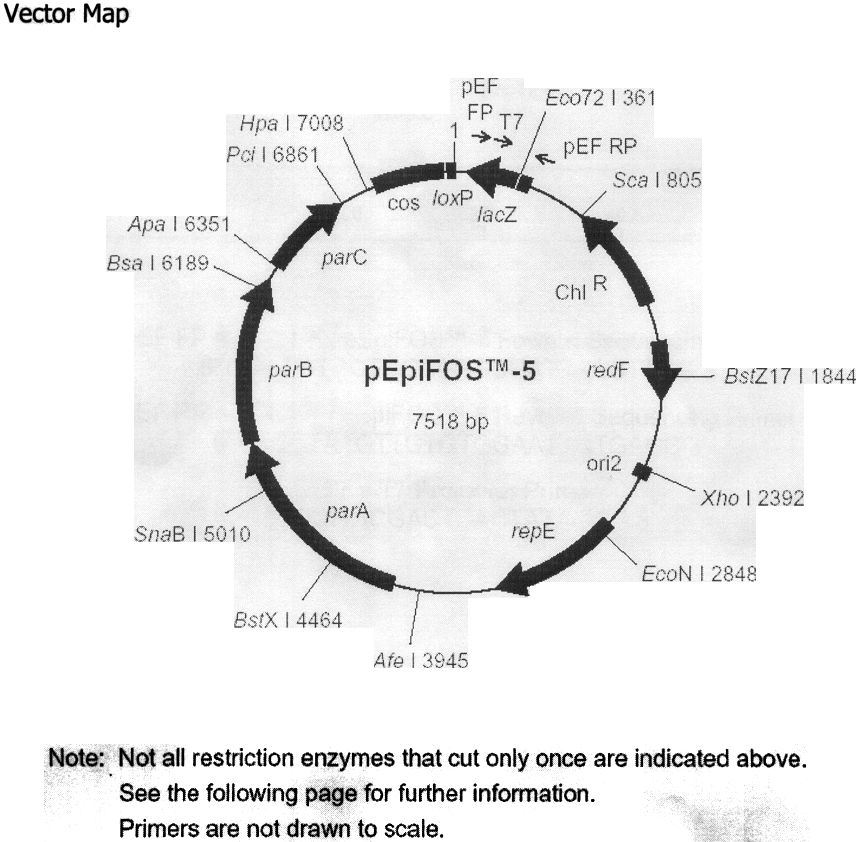
pEpiFOS-5
is a 7518 bp.
fosmid cloning vector which is provided Cloning-Ready -
linearized at the unique
Eco 72 I (blunt) restriction enzyme recognition site,
dephosphorylated and rigorously tested for purity and recombinant
cloning efficiency. Features of the vector include:
1)
Chloramphenicol-resistance as an antibiotic selectable marker.
2)
E. coli F
factor-based partitioning and copy number regulation system.
3)
Bacteriophage lambda cos site for lambda packaging
or lambda-terminase cleavage.
4)
Bacteriophage P1 Iox P site for Cre-recombinase cleavage.
5)
Bacteriophage T7 RNA polymerase promoter flanking the cloning
site.
6)
Fosmid end-sequencing
primers are available separately.
pEpiFOS-5
Sequencing Primers and Vector Data
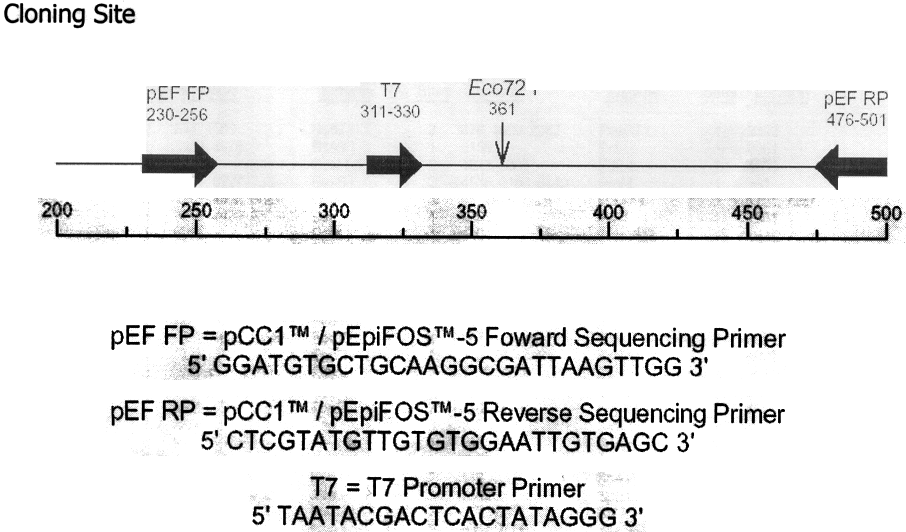
pCC1
/ pEpiFOS-5 Sequencing Primers
Primers
are available separately:
pCC1
TM I pEpiFOS
TM Forward Sequencing Primer Cat.
No. F5FPO10
5' GGATGTGCTGCAAGGCGATTAAGTTGG 3' ........ 1nmol supplied in TE Buffer at 50µM
pCC1
TM i pEpiFOS TM Reverse Sequencing Primer Cat.
No. F5RP011
5' CTCGTATGTTGTGTGGAATTGTGAGC 3' ................ 1nmol supplied in TE Buffer at 50µM
Note:
The sequence of the pCC1 / pEpiFOS Forward and Reverse Primers
do not function well as lRD800-labeled sequencing primers.
We recommend using the T7 and pCCl I pEpiFOS RP-2
Primers instead of the pCC1 / pEpiFOS Forward and Reverse
Primers respectively, for this purpose.
pCC1
TM / pEpiFOS TM RP-2 Reverse Sequencing Primer 5' TACGCCAAGCTATTTAGGTGAGA
3'

WIGR
Fosmid Prep
Growth
Media: 2XYT
Selection:
25ug/ml Chloramphenicol
Growth
Conditions: 14hr @ 37°C
The
prep is a simple alkaline lysis, followed by filtration through
a 0.45um cellulose acetate filter (Whatman) directly into
isopropanol. The pellet is washed twice with EtOH prior to
elution in water for sequencing.
|

