| Vector
pCYPAC2 Information |
 |

We have combined the best features of the P1 and BAC systems.
The pCYPAC2 vector has been constructed for the cloning of
large DNA fragments following electroporation. This
vector has been constructed by removing the stuffer fragment
form the pAd10SacBII vector (Pierce, J. C., Sauer, B. and
Sternberg, N. (1992) Proc. Natl. Acad. Sci, USA 89: 2056-2060.)
and inserting a pUC plasmid into the BamHI cloning site.
The pUC sequence has not been inserted in a permanent way.
During the cloning process, pUC sequences are removed through
a double-digestion scheme, which employs BamHI and ScaI.
The pUC plasmid cannot back into the BamHI cloning site, which
would otherwise result in the creation of non-recombinant
background clones in the library.
Re-ligation into the BamHI site is prevented at three levels:
1. pUC has been cleaved into ScaI fragments (ends incompatible
with BamHI)
2. The BamHI/ScaI oligo linkers have been physically
separated from the pUC and PAC-vector fragments and
3. All vector derived fragments have been treated with
alkaline phosphates to inhibit ligation of vector-to-vector
ends
pCYPAC2 clones have Kanamycin antibiotic resistance.
Clones should be grown in LB containing 25ug Kanamycin/ml.
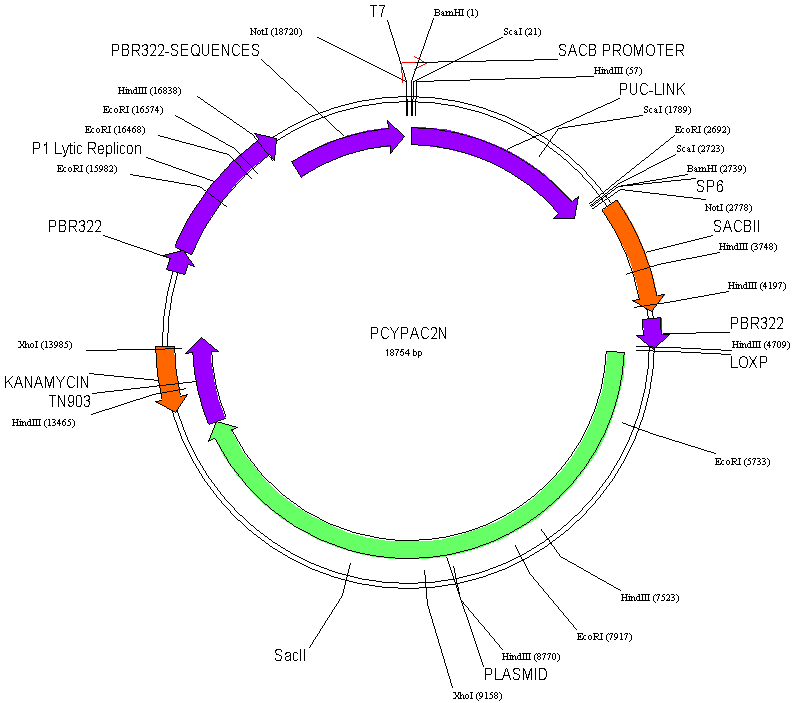
We have replaced the SciI site with a second
NotI site to allow release of most inserts as a single large
fragment. This facilitates size determination of inserts
as a single large fragment. This facilitates size determination
of inserts and determining the completeness of the NotI digest
reaction (via appearance of a constant vector product).
Contact BACPAC Resources ( [email protected]
) for further information regarding current availability
or ordering.
|

